Chapter 5 Welcome to the Tidyverse
“Data is like garbage. You’d better know what you are going to do with it before you collect it.”
- Mark Twain, 1835 - 1910
5.1 The Tidyverse
This chapter demonstrates the data management capabilities of the tidyverse (Wickham et al. 2019). Thus, Chapter 5 can be considered a tidyverse reconsideration of Ch 4. The tidyverse is currently a collection of eight core packages (Fig 5.1). These are:
- dplyr Grammar and functions for data manipulation.
- forcats Tools for solving common problems with factors.
- ggplot2 A system for “declaratively creating graphics”, based on the book The Grammar of Graphics (Wilkinson 2012).
- purrr An enhancement of R’s functional programming (FP) toolkit.
- readr Methods for reading rectangular data.
- stringr Functions to facilitate working with strings.
- tibble A “modern re-imagining of the data frame.”
- tidyr A set of functions for “tidying data.”

Figure 5.1: The main packages of the tidyverse.
The tidyverse library also contains several useful ancillary packages, including lubridate, reshape2, hms, blob, margrittr, and glue. While installing tidyverse will result in the installation of both main and ancillary packages, loading the tidyverse will result only in the complete loading of the eight main tidyverse packages.
Importantly, this chapter is not meant to be an authoritative summary of the tidyverse. Coverage here is mostly limited to the core data management packages magrittr, tibble, dplyr, stringer, and the ancillary packages lubridate and reshape2. The tidyverse ggplot2 package is the major focus of Chapter 7. Wickham, Çetinkaya-Rundel, and Grolemund (2023) provides a succinct but thorough introduction to the tidyverse in the open source book R for Data Science. Useful tidyverse “cheatsheets” can be found here.
The tidyverse packages can be downloaded using:
5.2 Pipes
An important convention of the tidyverse is the widespread use of the forward pipe operator: |> . In programming, a pipe is a set of commands connected in series, where the output of one command is the input of the next1.
In many cases, use of pipes allows clearer representations of coding processes2. Incidentally, the |> pipe, from the base package, is motivated by an older forward pipe operator from the tidyverse package magrittr, %>%. As of R 4.1, the native pipe operator for the tidyverse is |> (although %>% will still work if magrittr is loaded)3. Notably, while |> is more syntactically (and algorithmically) streamlined than %>%, there are several features available to %>% that do not exist for |>, including the potential for a placeholder operator4. Nonetheless, I focus on |>, not %>%, here.
Example 5.1 \(\text{}\)
Consider the circular operation: \(\log_e(\exp(1))\). We could write this as,
[1] 1Here the number \(1\) is piped into the function exp(), with the result: \(\exp(1) = e^1 = e\), and this outcome is piped into the function log(), with the result: \(\log_e e = 1\). Because the first arguments of exp() and log() are simply calls to numeric data, and these are provided by the previous pipe segment, we do not have to include information about x for f(x) operations. Thus, when functions require only the previous pipe segment result as an argument, then x |> f() is equivalent to \(f(x)\)5. In the case that multiple arguments need to be specified, the script x |> f(y) is equivalent to \(f(x, y)\), and x |> f(y) |> g(z) describes \(g(f(x, y), z)\). For instance,
[1] 3.321928\(\blacksquare\)
Example 5.2 \(\text{}\)
This example illustrates that the forward pipe works recursively from the result of the previous pipe segment.
height age Seed
1 4.51 3 301
15 10.89 5 301
29 28.72 10 301
43 41.74 15 301
57 52.70 20 301
71 60.92 25 301 height age Seed
57 52.70 20 301
71 60.92 25 301\(\blacksquare\)
Example 5.3 \(\text{}\)
We can define the result of a pipe to be a global variable. Consider the script below (Fig 5.2).
x <- seq(1,10,length=100)
y <- x |> sin()
plot(x, y, type = "l", ylab = "sin(x)", xlab = "x (radians)")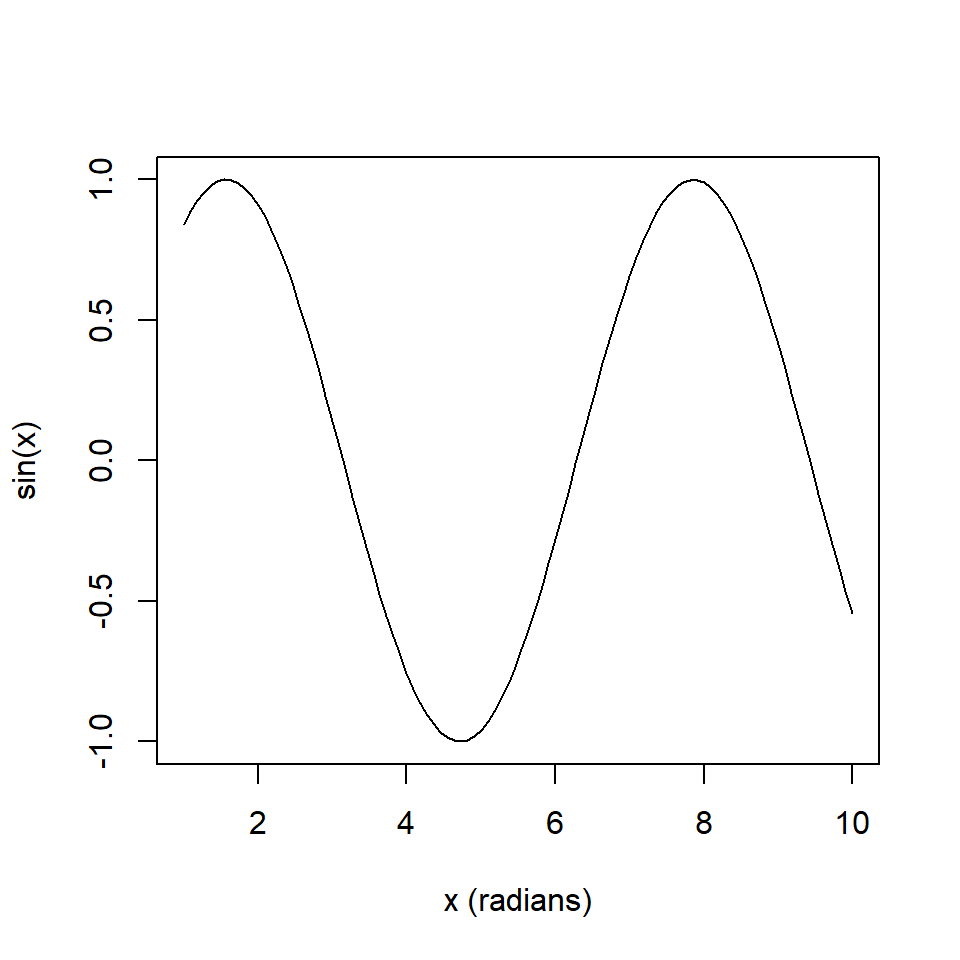
Figure 5.2: Creating a global variable (object) resulting from a pipeline.
\(\blacksquare\)
5.2.1 Other Pipes
It is worth noting that, in addition to %>%, magrittr contains several other potentially useful pipe operators. These include the assignment pipe and the tee pipe. The assignment pipe operator, %<>%, will pipe x into one or more f(x) expressions, and then assign the result to the name x6. The tee pipe operator %T>% works like %>%, except the return value in x %T>% f(x) is x itself. This is useful when a pipeline requires a side-effect like plotting or printing7.
5.3 tibble
The tidyverse package tibble provides an alternative to the data.frame format of data storage, called a tibble. Tibbles have classes dataframe and tbl_df, allowing them to posses additional characteristics including enhanced printing (see Example 5.4 below).
Additional distinguishing characteristics of tibbles include: 1) a character vector is not automatically coerced to have class factor, 2) recycling (see Section 3.1.1) only occurs for an input of length one, and 3) there is no partial matching when $ is used to index by tibble columns by name8. The functions tibble() generates tibbles. The function as_tibble() coerces a dataframe to be a tibble.
Example 5.4 \(\text{}\)
Here we compare dataframe and tibble output of the same data.
data <- data.frame(numbers = 1:3, letters = c("a","b","c"),
date = as.Date(c("2021-12-1", "2021-12-2",
"2021-12-2"),
format = "%Y-%m-%d"))
data numbers letters date
1 1 a 2021-12-01
2 2 b 2021-12-02
3 3 c 2021-12-02# A tibble: 3 x 3
numbers letters date
<int> <chr> <date>
1 1 a 2021-12-01
2 2 b 2021-12-02
3 3 c 2021-12-02\(\blacksquare\)
5.4 dplyr
The dplyr package contains a collection of core tidyverse algorithms for data manipulation9. Table 5.1 lists some useful dplyr functions.
| Function | Usage |
|---|---|
summarise() |
Numerical summaries of variables. |
group_by |
Group a dataframe by a categorical variable. |
filter() |
Subset variables based on outcomes. |
arrange() |
Reorder rows in a dataframe or tibble. |
mutate() |
Creates new variables from functions of existing variables. |
select() |
Selects variables from tibbles or dataframes. |
5.4.1 summarize()
The function summarize(), or equivalently summarise(), creates a new data frame with one row for each combination of specified grouping variables. If no groups are given (for instance, in the case that group_by is not used to group data), dataframe rows will be summaries of all observations in the required input .data argument.
Example 5.5 \(\text{}\)
Here we use summarize() to obtain means for loblolly pine height (in feet) and age (in years).
mean.height.ft mean.age.yrs
1 32.3644 13\(\blacksquare\)
5.4.2 group_by()
The group_by() function is often used in conjunction with other dplyr functions, including summarize(), to provide an underlying grouping framework for data summaries.
Example 5.6 \(\text{}\)
Here we use group_by() with summarize() to describe the Loblolly height data. Specifically, we will take the mean and the variance of Loblolly$height with respect to categories specified in group_by().
Loblolly |>
group_by(Seed) |>
summarise(mean.height.ft = mean(height),
var.height.ft2 = var(height)
) |>
head(5)# A tibble: 5 x 3
Seed mean.height.ft var.height.ft2
<ord> <dbl> <dbl>
1 329 30.3 443.
2 327 30.6 440.
3 325 31.9 468.
4 307 31.3 494.
5 331 31.0 495.More than only grouping variable can be specified in group_by():
Loblolly |>
group_by(Seed, age) |>
summarise(mean.height.ft = mean(height),
var.height.ft2 = var(height)
) |>
head(5)`summarise()` has grouped output by 'Seed'. You can override using the
`.groups` argument.# A tibble: 5 x 4
# Groups: Seed [1]
Seed age mean.height.ft var.height.ft2
<ord> <dbl> <dbl> <dbl>
1 329 3 3.93 NA
2 329 5 9.34 NA
3 329 10 26.1 NA
4 329 15 37.8 NA
5 329 20 48.3 NAClearly, group_by() and summarise() allow more options than the base function tapply() (Section 4.1.1.4). The latter function only provides summaries of groups within a single categorical INDEX, with respect to a single quantitative vector, and a single user-defined function.
Starting with dplyr 1.1.0, we can use the .by argument in summarize to bypass group_by(), although this argument is experimental, and may be deprecated in the future (see ?summarise).
Loblolly |>
summarise(mean.height.ft = mean(height),
var.height.ft2 = var(height),
.by = Seed) |>
head(5) Seed mean.height.ft var.height.ft2
1 301 33.24667 512.4979
2 303 34.10667 552.2362
3 305 35.11500 572.5056
4 307 31.32833 493.8291
5 309 33.78167 535.1160\(\blacksquare\)
5.4.3 filter()
The function filter() provides a straightforward way to extract dataframe rows based on Boolean operators.
Example 5.7 \(\text{}\)
Here we obtain rows in Loblolly associated with seed type 301.
height age Seed
1 4.51 3 301
15 10.89 5 301
29 28.72 10 301
43 41.74 15 301
57 52.70 20 301
71 60.92 25 301Here are Loblolly rows associated with height responses greater than 60 feet.
height age Seed
71 60.92 25 301
72 63.39 25 303
73 64.10 25 305
75 63.05 25 309
77 60.07 25 315
78 60.69 25 319
79 60.28 25 321
80 61.62 25 323\(\blacksquare\)
5.4.4 arrange()
The function arrange() orders the rows of a data frame based on the alphanumeric ordering of specified data.
Example 5.8 \(\text{}\)
Here we use arrange() to sort the result from the previous chunk from smallest to largest loblolly pine heights.
height age Seed
77 60.07 25 315
79 60.28 25 321
78 60.69 25 319
71 60.92 25 301
80 61.62 25 323
75 63.05 25 309
72 63.39 25 303
73 64.10 25 305One can use arrange(desc()) to sort a dataframe in descending (largest-to-smallest) order.
height age Seed
73 64.10 25 305
72 63.39 25 303
75 63.05 25 309
80 61.62 25 323
71 60.92 25 301
78 60.69 25 319
79 60.28 25 321
77 60.07 25 315\(\blacksquare\)
5.4.5 slice_min() and slice_max()
The helpful dplyr functions slice_min() and slice_max() allow subsetting of dataframe rows by minimum and maximum values in some column, respectively.
Example 5.9 \(\text{}\)
height age Seed
73 64.10 25 305
72 63.39 25 303
75 63.05 25 309
80 61.62 25 323
71 60.92 25 301\(\blacksquare\)
5.4.6 select()
The select() function allows one to select particular variables in a data frame.
Example 5.10 \(\text{}\)
For instance, here I select height from Loblolly.
height
1 4.51
15 10.89
29 28.72
43 41.74
57 52.70
71 60.92\(\blacksquare\)
The select() function can be used in more sophisticated ways by combining it with other dplyr functions like starts_with() and ends_with(), or other Boolean operators.
Example 5.11 \(\text{}\)
He we select the height and age columns by calling for variable names that start with "h" or end with "e".
height age
1 4.51 3
15 10.89 5
29 28.72 10\(\blacksquare\)
5.4.7 mutate()
The function mutate() creates new dataframe columns that are functions of existing variables.
Example 5.12 \(\text{}\)
Below we select the age and height columns using select(), convert height in feet to height in meters using mutate(), plot the result as a side-task using the tee pipe, %T>% (note the use of the . placeholder operator) (Fig 5.3), and then take the column means of age and height. Note that by default, all columns from the previous pipe segment will be in the mutate() output although all columns need not be explicitly mutated. Output columns can be specified using the mutate() argument .keep.
library(magrittr) # to access tee pipe
Loblolly |>
select(c(age, height)) |>
mutate(height = height * 0.3048) %T>%
plot(.,ylab = "Height (m)", xlab = "Age (yrs)") |>
colMeans() age height
13.000000 9.864671 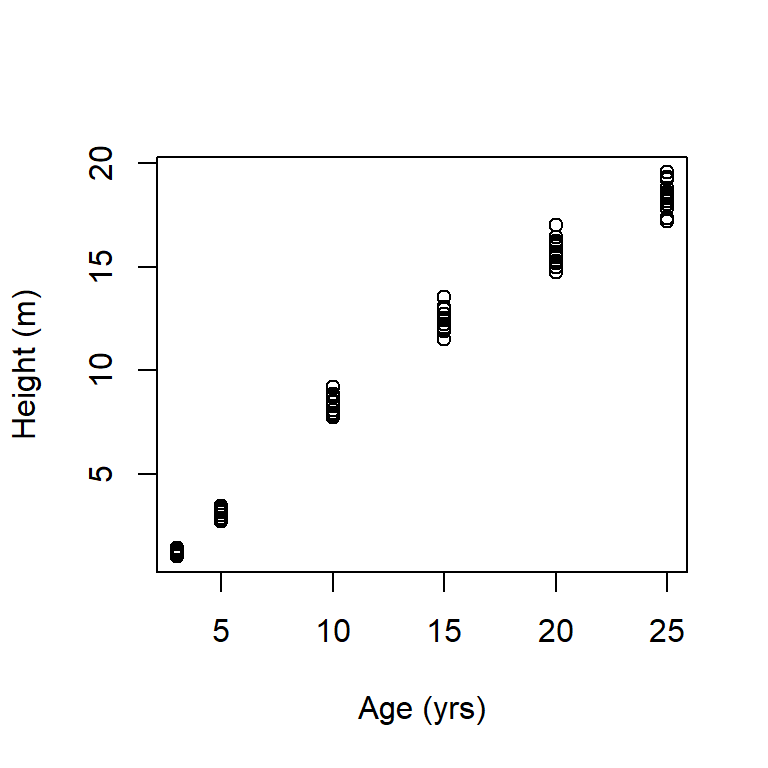
Figure 5.3: Plot of loblolly pine height as a function of age, after converting height to meters. In base R dialect we could use: with(Loblolly, plot(age, height * 0.3048, ylab = "Height (m)", xlab = "Age (yrs)")). This is quite a bit harder to decipher.
\(\blacksquare\)
5.4.8 across()
The dplyr function across() allows extensions similar to those in apply() wherein the same function can be applied to all columns in the first argument of across(). Specifying the first argument in across() as everything() would allow application of a function to all columns in a dataframe.
Example 5.13 \(\text{}\)
Here we take the medians of the quantitative columns in Loblolly using across() and summarize().
age height
1 12.5 34\(\blacksquare\)
5.5 stringr
As evident in Section 4.3, use of regular expressions for matching, querying and substituting strings can be confusing. The stringr package attempts to simplify some of these difficulties.
The stringr package uses processing tools from the package stringi (Gagolewski 2022) for pattern searching under a wide array of potential approaches. All stringr functions have the prefix str_ and take a character string vector as the first argument.
Consider the vector of plant scientific names used to demonstrate string management in Section 4.3.
names = c("Achillea millefolium", "Aster foliaceus",
"Elymus scribneri", "Erigeron rydbergii",
"Carex elynoides", "Carex paysonis",
"Taraxacum ceratophorum")Example 5.14 \(\text{}\)
The function str_length() can be used to count the number of characters in a string.
[1] 20 15 16 18 15 14 22\(\blacksquare\)
Example 5.15 \(\text{}\)
The function str_detect() tests for the presence or absence of a pattern in a string. Here I test for presence of the genus Aster.
[1] FALSE TRUE FALSE FALSE FALSE FALSE FALSEHere are entries not containing Aster.
[1] TRUE FALSE TRUE TRUE TRUE TRUE TRUE\(\blacksquare\)
Example 5.16 \(\text{}\)
Here we subset names using the function stringr::str_subset() to obtain species within the genus Carex.
[1] "Carex elynoides" "Carex paysonis" \(\blacksquare\)
Example 5.17 \(\text{}\)
The function str_replace() is analogous to the base R function gsub(). It can be used to replace text based on a pattern.
[1] "Achillea millefolium" "Aster foliaceus"
[3] "Elymus scribneri" "Erigeron rydbergii"
[5] "C. elynoides" "C. paysonis"
[7] "Taraxacum ceratophorum"\(\blacksquare\)
Most stringr functions work with regular expressions (Section 4.3.6).
Example 5.18 \(\text{}\)
Here we count upper and lower case vowels with the function stringr::str_count() using a pattern defined by the regex character class [AEIOUaeiou].
[1] 9 7 5 7 6 5 9and use stringr::str_extract() to extract strings nine alphanumeric characters long, and then sort the strings with a pipe.
[1] "elynoides" "foliaceus" "millefoli" "rydbergii" "scribneri" "Taraxacum"\(\blacksquare\)
5.6 lubridate
Base R approaches for handling date-time data are described in Section 4.4. The package lubridate (https://lubridate.tidyverse.org/) contains functions for simplifying and extending some of these operations.
Example 5.19 \(\text{}\)
As an example dataset, I will use the time series used to illustrate date-time classes in Section 4.4.
dates <- c("08/13/2019 04:00", "08/13/2019 06:30", "08/13/2019 09:00",
"08/13/2019 11:30", "08/13/2019 14:00", "08/13/2019 16:30",
"08/13/2019 19:00", "08/13/2019 21:30", "08/14/2019 00:00",
"08/14/2019 02:30", "08/14/2019 05:00", "08/14/2019 07:30",
"08/14/2019 10:00", "08/14/2019 12:30", "08/14/2019 15:00",
"08/14/2019 17:30", "08/14/2019 20:00", "08/14/2019 22:30",
"08/15/2019 01:00", "08/15/2019 03:30")
library(lubridate)We will define the timezone to be timezone of our computer workstation.
The package lubridate contains data-time parsers that may be easier to use than the base functions strptime and as.Date.
For the current example, we note that the data are in a month/day/year hour:minute format. So we can create a time series using the function lubridate::mdy_hm.
[1] "2019-08-13 04:00:00 MDT" "2019-08-13 06:30:00 MDT"
[3] "2019-08-13 09:00:00 MDT" "2019-08-13 11:30:00 MDT"
[5] "2019-08-13 14:00:00 MDT" "2019-08-13 16:30:00 MDT"
[7] "2019-08-13 19:00:00 MDT" "2019-08-13 21:30:00 MDT"
[9] "2019-08-14 00:00:00 MDT" "2019-08-14 02:30:00 MDT"
[11] "2019-08-14 05:00:00 MDT" "2019-08-14 07:30:00 MDT"
[13] "2019-08-14 10:00:00 MDT" "2019-08-14 12:30:00 MDT"
[15] "2019-08-14 15:00:00 MDT" "2019-08-14 17:30:00 MDT"
[17] "2019-08-14 20:00:00 MDT" "2019-08-14 22:30:00 MDT"
[19] "2019-08-15 01:00:00 MDT" "2019-08-15 03:30:00 MDT"Other lubridate parsers include ymd(), ymd_hms(), dmy(), dmy_hms(), and mdy(). The lubridate parsers can often handle mixed methods of data entry. From the ymd() documentation we have the following example:
x <- c(20090101, "2009-01-02", "2009 01 03", "2009-1-4",
"2009-1, 5", "Created on 2009 1 6", "200901 !!! 07")
ymd(x)[1] "2009-01-01" "2009-01-02" "2009-01-03" "2009-01-04" "2009-01-05"
[6] "2009-01-06" "2009-01-07"\(\blacksquare\)
Lubridate also allows extended mathematical operations for its date-time objects with the functions duration(), period(), and interval().
Example 5.20 \(\text{}\)
Duration functions include dseconds(), dminutes(), ddays(), and dmonths().
[1] "31557600s (~1 years)"[1] "31557600s (~1 years)"[1] "2019-08-13 04:00:00 MDT"[1] "2019-08-14 04:00:00 MDT"\(\blacksquare\)
Example 5.21 \(\text{}\)
Periodic functions include seconds(), minutes(), hours(), and days().
[1] "12d 0H 2M 3S"[1] "2019-08-13 04:00:00 MDT"[1] "2019-08-01 04:00:00 MDT"\(\blacksquare\)
Example 5.22 \(\text{}\)
Interval functions include int_length(), int_start(), and int_end().
[1] "171000s (~1.98 days)"\(\blacksquare\)
5.7 reshape2
Tidyverse functions generally require that data are in a long table format. That is, data are stored with columns containing all the values for a particular variable of interest. Unfortunately, this format is not conventional for many scientific applications, particularly longitudinal studies that follow experimental units over time. These will often have a wide table format. The tidyverse reshape2 package contains several functions for converting dataframes from wide to a long table formats, including the functions reshape2::melt() and tidyr::gather(). The reshape2::melt.data.frame() function generates a value column based on data commonalities of outcomes given in a variable or variables defined in the id argument. A remaining column, if any, that captures these commonalities will be given the name variable. The names of the value and variable output columns can be changed with the arguments value.name and variable.name, respectively.
Example 5.23 \(\text{}\)
Consider the asbio::asthma dataset, which has a wide table format.
The dataset documents the effect of three respiratory treatments (measured as Forced Expiratory Volume in one second (FEV1))
for 24 asthmatic patients over time (11H - 18H, i.e, hour 11 to hour 18). A baseline measure of FEV1 (BASEFEV1) was also taken 11 hours before application of the treatment.
PATIENT BASEFEV1 FEV11H FEV12H FEV13H FEV14H FEV15H FEV16H FEV17H FEV18H
1 201 2.46 2.68 2.76 2.50 2.30 2.14 2.40 2.33 2.20
2 202 3.50 3.95 3.65 2.93 2.53 3.04 3.37 3.14 2.62
3 203 1.96 2.28 2.34 2.29 2.43 2.06 2.18 2.28 2.29
4 204 3.44 4.08 3.87 3.79 3.30 3.80 3.24 2.98 2.91
5 205 2.80 4.09 3.90 3.54 3.35 3.15 3.23 3.46 3.27
6 206 2.36 3.79 3.97 3.78 3.69 3.31 2.83 2.72 3.00
DRUG
1 a
2 a
3 a
4 a
5 a
6 alibrary(reshape2)
asthma.long <- asthma |> melt(id = c("DRUG", "PATIENT"),
value.name = "FEV1",
variable.name = "TIME")
# here I simplify the names in the TIME variable
asthma.long$TIME <- factor(asthma.long$TIME,
labels = c("BASE",
paste("H", 11:18, sep = "")))
head(asthma.long) DRUG PATIENT TIME FEV1
1 a 201 BASE 2.46
2 a 202 BASE 3.50
3 a 203 BASE 1.96
4 a 204 BASE 3.44
5 a 205 BASE 2.80
6 a 206 BASE 2.36In the code above, the function reshape2::melt() is used to convert to a long table format, and time designations are simplified using the base function factor(). The factor() function can be used to create a categorical variable with particular levels (Section 3.3), or to change the names of levels. The latter application is used here.
\(\blacksquare\)
Exercises
- Create a tibble from the
Downsdataframe shown below. The data comprise part of a report summarizing Down’s syndrome cases in British Columbia, compiled by the British Columbia Health Surveillance Registry (Geyer 1991).Examine both the original
Downsdataframe and the tibble representation ofDownsby printing them. Do we gain additional information from the tibble?Find the mean and variance of the
Agecolumn from theDownsdataset using pipes and dpylr functions.
- Bring in the
world.emissionsdataset from package asbio.- Using the forward pipe operator,
|>, andfilter()from dplyr, create a dataframe of just US data. - Using
|>,filter(), andsummarise(), find the first and last year of emissions data for the US. - Using
|>,%T>%,filter(),mutate(), andplot(), plot per capita CO\(_2\) emissions for the US by year (as an intermediate pipeline step) and find the maximum CO\(_2\) emission level. Hint: see Exercise 5.12. - Using
|>andfilter()create a new dataframe calledno.repeatsthat eliminates rows with the entry"redundant"in theworld.emissions$continentcolumn. - With the
no.repeatsdataframe and the functionsgroup_by(), andsummarise(), get mean CO\(_2\) levels for each country over time. - Using
|>,group_by(),summarise()andslice_max(), identify the 10 countries with the highest recorded cumulative CO\(_2\) emissions.
- Using the forward pipe operator,
- Consider the character vector
omicsbelow (Bonnin 2021).Use
stringr::str_detect()to test for strings with the pattern"genom".Using
str_detect(), test for strings starting with the pattern"genom"by using an extended regular expression:^genomin thestr_detect()argumentpattern(see Section 4.3.6.1).Using
str_detect(), test for strings ending with the pattern"omics"by using an extended regular expression (see Section 4.3.6.1).Using
str_subset(), subset the string vectoromicsto string entries containing the pattern"genom".Using
str_replace(), replace the text"omics"with"ome".
- Consider the character vector
timesbelow, which has the format:day-month-year hour:minute:second.Convert
timesinto a lubridate date-time object using an appropriate lubridate function.Add two days and seven seconds to each entry in
timeusinglubridate::days.Using lubridate functions, find the difference, in seconds, between the beginning and the end of the time series.
References
Pipe programming dates back to early developments in Unix operating systems (Ritchie 1984; Bell Labs 2004), wherein pipes are codified as vertical bars
"|". Along with Unix/Linux, pipes are widely used in the languages F#, Julia, and JavaScript, among others. ↩︎In particular, when you see
|>it is helpful to think “and then”.↩︎The RStudio shortcut for
%>%is Ctrl\(+\)Shift\(+\)m. To force RStudio to default to|>when using Ctrl\(+\)Shift\(+\)m (or some other keyboard shortcut), one can modify appropriate settings in Tools\(>\)Global Options\(>\)Code.↩︎In general, the dot placeholder operator,
., from magrittr allows operations like \(f(x,y)\) by specifyingx |> f(.,y). For example:2 %>% log(10, base = .). In this script the number 2 will be piped into thebaseargument in the functionlog().↩︎The
%>%forward pipe does not even require the()no argument designation. That is,x %>% fis equivalent to \(f(x)\).↩︎For instance,
library(magrittr); x <- -4:4; x %<>% abs %>% sort; xwould print the pipe-modified version ofx.↩︎For instance,
rnorm(20) |> matrix(ncol = 2) %T>% plot |> colSums. In this case a plot and the sums of columns will both be printed (see Example 5.12).↩︎According to package tibble: “\(\ldots\)tibbles are lazy and surly: they do less and complain more than base dataframes. This forces problems to be tackled earlier and more explicitly, typically leading to code that is more expressive and robust.”↩︎
dplyr has largely replaced the now retired plyr package.↩︎